Analyzing DNA
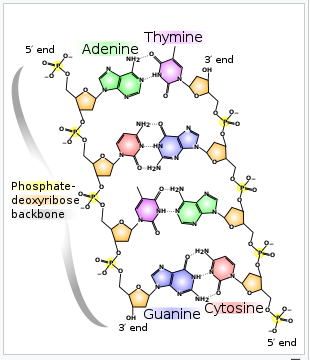
DNA is a molecule composed of two polynucleotide chains wrapped around each other in the shape of a double helix.
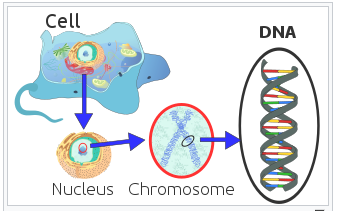
DNA is further organized into chromosomes which are packed into the nucleus of a cell.
So how do we get data from a nucleotide polymer in a chromosome, in a nucleus, in a cell? Well a common approach is to leverage something called Next Generation Sequencing. Here is how that is accomplished:
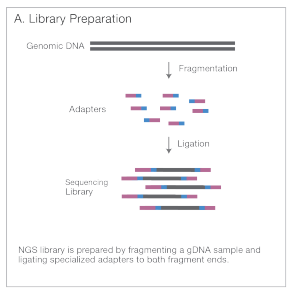
- Library Preparation: DNA is fragmented, adapters are added to those fragments - creating a Sequencing library.
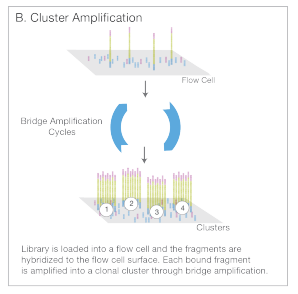
- Cluster Amplification: The adapters then are stuck to a flow cell, and these bound fragments are amplified to increase the signal.
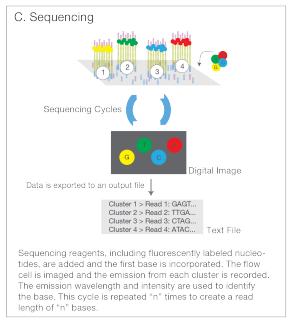
- Sequencing: Fluorescently labeled nucleotides are added to the flow cell and as they attach to the DNA they emit a signal which is picked up by the sequencing machine, telling it the identity and position of the base in the fragment. These fragments are then read to create reads - text files with sequence of each dna fragment.
- Alignment/Data Analysis: Now that we have these reads, we can align them to a known sequence and perform analyses.
With these reads we can perform all kinds of analysis workflows. Here we explore the following:
References
- An Introduction to Next-Generation Sequencing Technology _____________________________________________________________
Next Workshop: Assembly